Example
Diabetes in Pima Indian Women
Characteristics of Pima Indian women tested for diabetes are used in this example to predict their disease statuses.
| Characteristic | Value |
|---|---|
| Number of women | 392 |
| diabetes | |
| pos | 130 (33.16%) |
| neg | 262 (66.84%) |
| pregnant | |
| Median (Range) | 2 (0, 17) |
| glucose | |
| Median (Range) | 119 (56, 198) |
| pressure | |
| Median (Range) | 70 (24, 110) |
| triceps | |
| Median (Range) | 29 (7, 63) |
| insulin | |
| Median (Range) | 125.5 (14, 846) |
| mass | |
| Median (Range) | 33.2 (18.2, 67.1) |
| pedigree | |
| Median (Range) | 0.4495 (0.085, 2.420) |
| age | |
| Median (Range) | 27 (21, 81) |
Training Set Analysis
## Analysis libraries
library(MachineShop)
library(ggplot2)
## Parallel processing
library(doParallel)
registerDoParallel(cores = 6)
## Dataset
data(PimaIndiansDiabetes2, package = "mlbench")
Pima <- na.omit(PimaIndiansDiabetes2)
## Model formula
fo <- diabetes ~ .
## Model selected from tuned models
selected_model <- SelectedModel(TunedModel(KNNModel, grid = 5),
TunedModel(NNetModel, grid = 5),
TunedModel(RandomForestModel, grid = 5))
## Model fit
model_fit <- fit(fo, data = Pima, model = selected_model)
## Variable importance
vi <- varimp(model_fit)
plot(vi)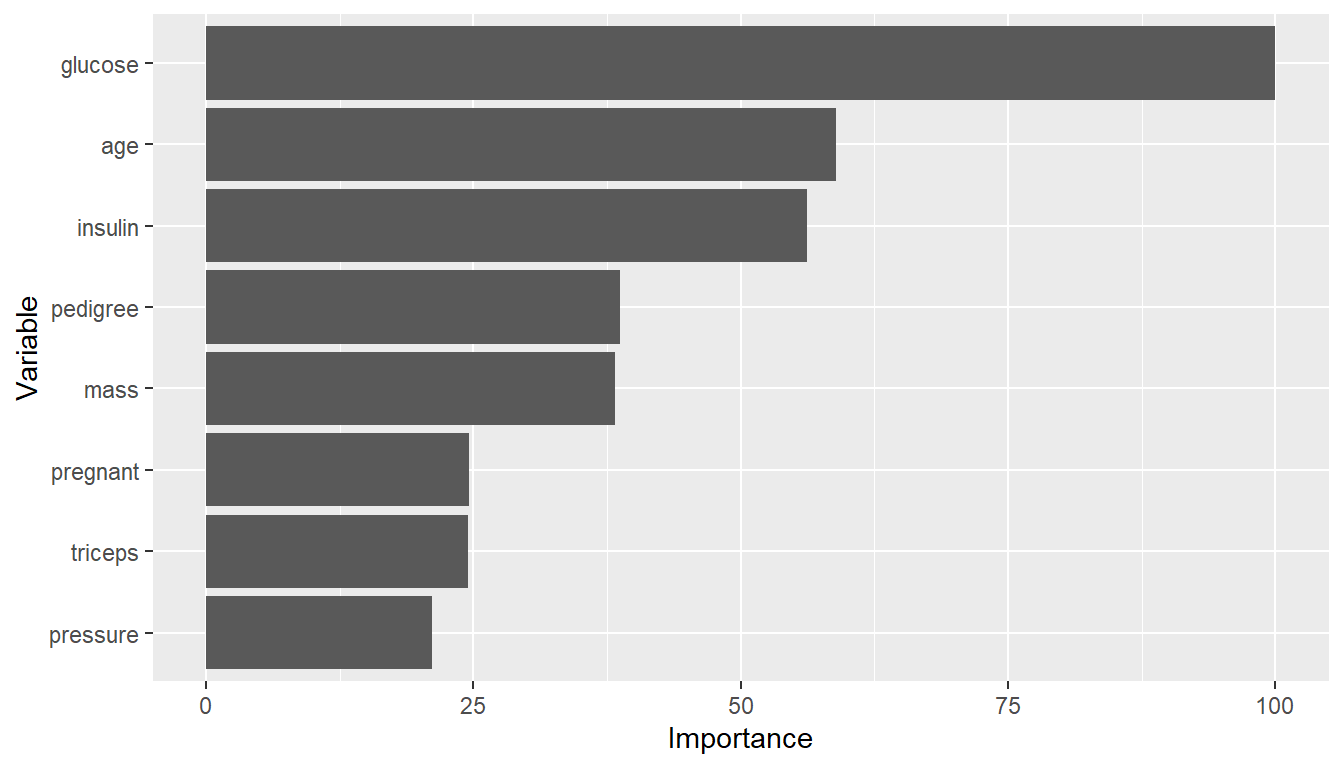
Generalization Performance
## Resample estimation with cross-validation
res <- resample(fo, data = Pima, model = selected_model, control = CVControl)
## Estimated performance
summary(performance(res))
#> Statistic
#> Metric Mean Median SD Min Max NA
#> Brier 0.1441490 0.1385509 0.04313255 0.1060426 0.2562013 0
#> Accuracy 0.7857051 0.7974359 0.06529521 0.6410256 0.8717949 0
#> Kappa 0.4999248 0.5123220 0.15928724 0.1250000 0.6808511 0
#> ROC AUC 0.8531339 0.8786982 0.08404156 0.6375740 0.9186391 0
#> Sensitivity 0.6153846 0.6153846 0.13074409 0.3076923 0.7692308 0
#> Specificity 0.8700855 0.8682336 0.06074830 0.8076923 1.0000000 0
## Variable probability cutoff
summary(performance(res, cutoff = 0.25))
#> Statistic
#> Metric Mean Median SD Min Max NA
#> Brier 0.1441490 0.1385509 0.04313255 0.1060426 0.2562013 0
#> Accuracy 0.7499359 0.7435897 0.06878381 0.6410256 0.8500000 0
#> Kappa 0.5013070 0.5080645 0.14027061 0.2500000 0.6946565 0
#> ROC AUC 0.8531339 0.8786982 0.08404156 0.6375740 0.9186391 0
#> Sensitivity 0.8846154 0.9230769 0.12162606 0.6153846 1.0000000 0
#> Specificity 0.6831909 0.6730769 0.06161579 0.5925926 0.7777778 0ROC Curve
## True positive and false positive rates over all probability cutoffs
roc <- performance_curve(res)
## ROC curve
plot(roc, diagonal = TRUE) + coord_fixed()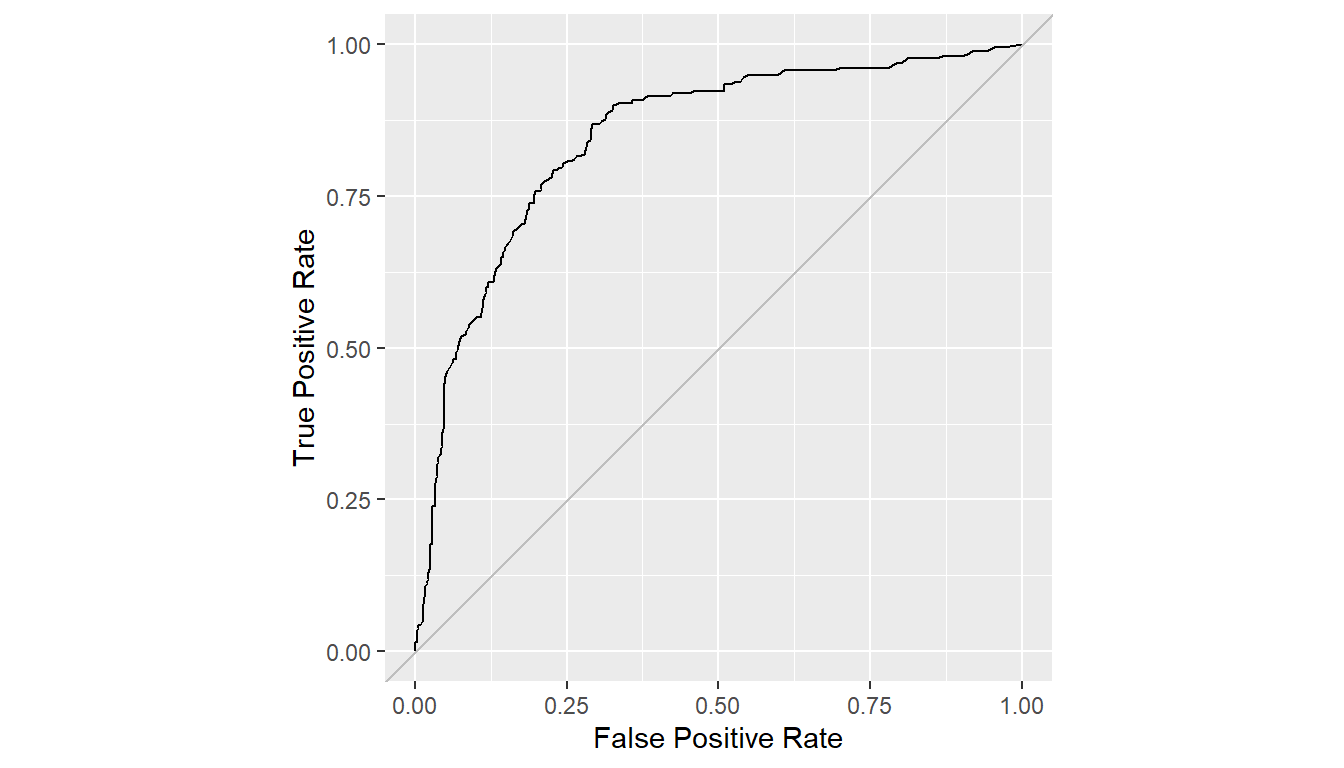
## Area under the curve
auc(roc)
#> Model: SelectedModel
#> [1] 0.8464381Confusion Matrices
(conf <- confusion(res))
#> --- ConfusionList object ----------------------------------------------------
#>
#> === $SelectedModel ==========================================================
#> === BinaryConfusionMatrix object ===
#> Observed
#> Predicted neg pos
#> neg 228 50
#> pos 34 80
summary(conf)
#> --- $SelectedModel ----------------------------------------------------------
#> Number of responses: 392
#> Accuracy (SE): 0.7857143 (0.02072459)
#> Majority class: 0.6683673
#> Kappa: 0.5011514
#>
#> neg pos
#> Observed 0.6683673 0.3316327
#> Predicted 0.7091837 0.2908163
#> Agreement 0.5816327 0.2040816
#> Sensitivity 0.8702290 0.6153846
#> Specificity 0.6153846 0.8702290
#> PPV 0.8201439 0.7017544
#> NPV 0.7017544 0.8201439
plot(conf)
#> $SelectedModel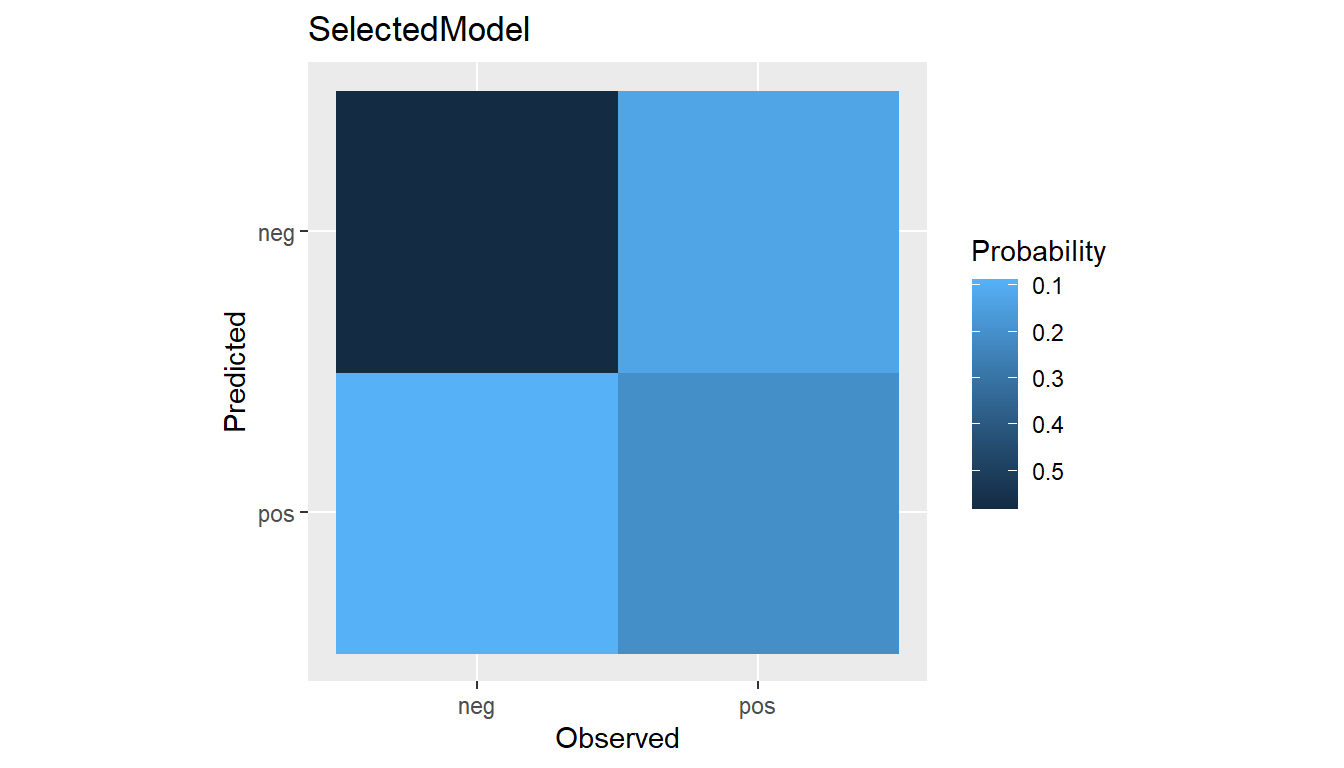
## Variable probability cutoff
summary(confusion(res, cutoff = 0.25))
#> --- $SelectedModel ----------------------------------------------------------
#> Number of responses: 392
#> Accuracy (SE): 0.75 (0.02187044)
#> Majority class: 0.6683673
#> Kappa: 0.5017122
#>
#> neg pos
#> Observed 0.6683673 0.3316327
#> Predicted 0.4948980 0.5051020
#> Agreement 0.4566327 0.2933673
#> Sensitivity 0.6832061 0.8846154
#> Specificity 0.8846154 0.6832061
#> PPV 0.9226804 0.5808081
#> NPV 0.5808081 0.9226804Calibration Curve
cal <- calibration(res, breaks = NULL)
plot(cal, se = TRUE)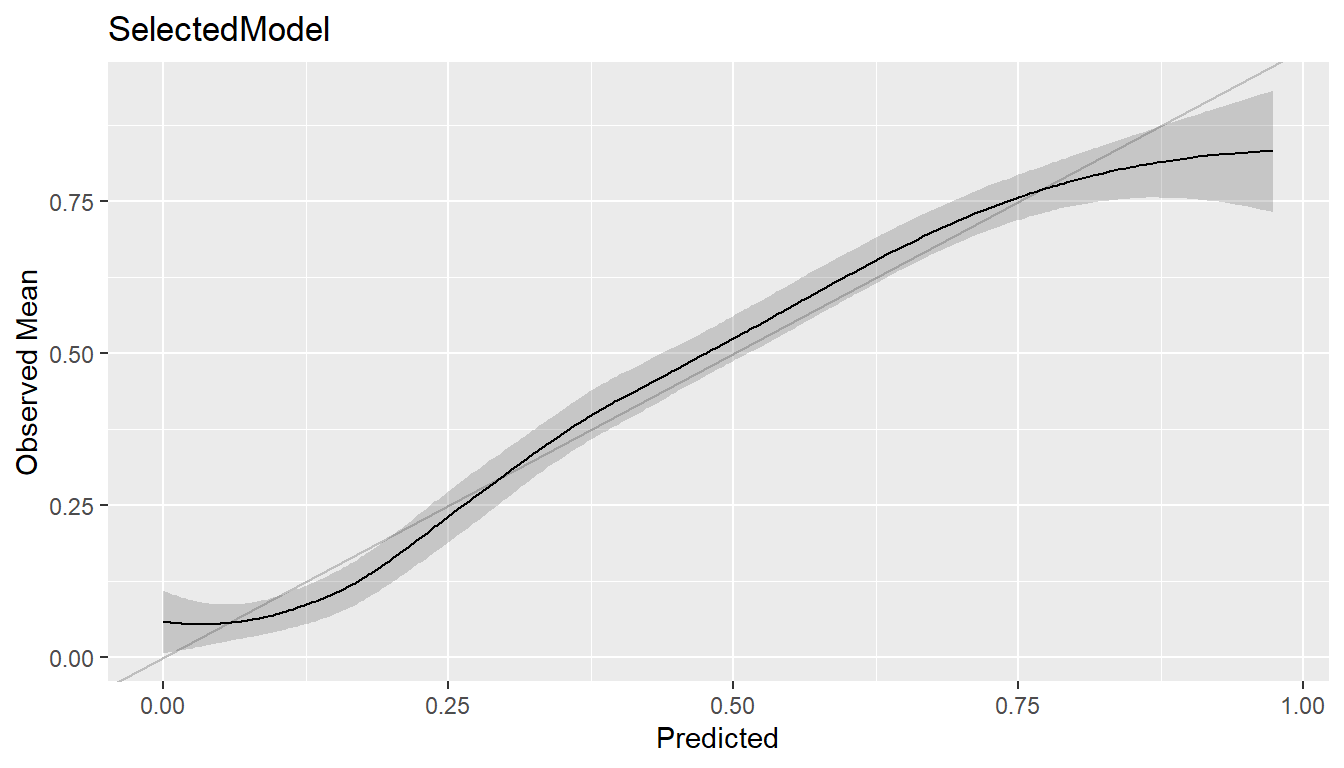
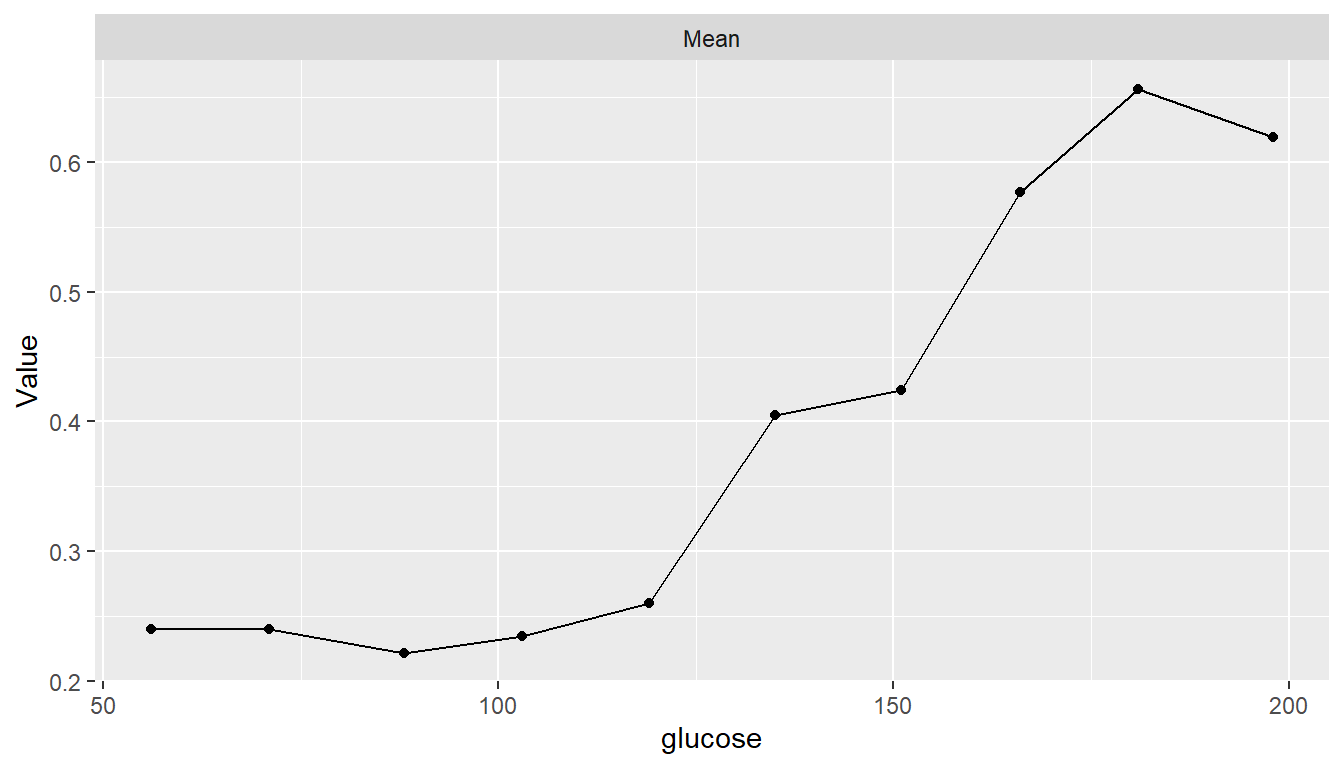
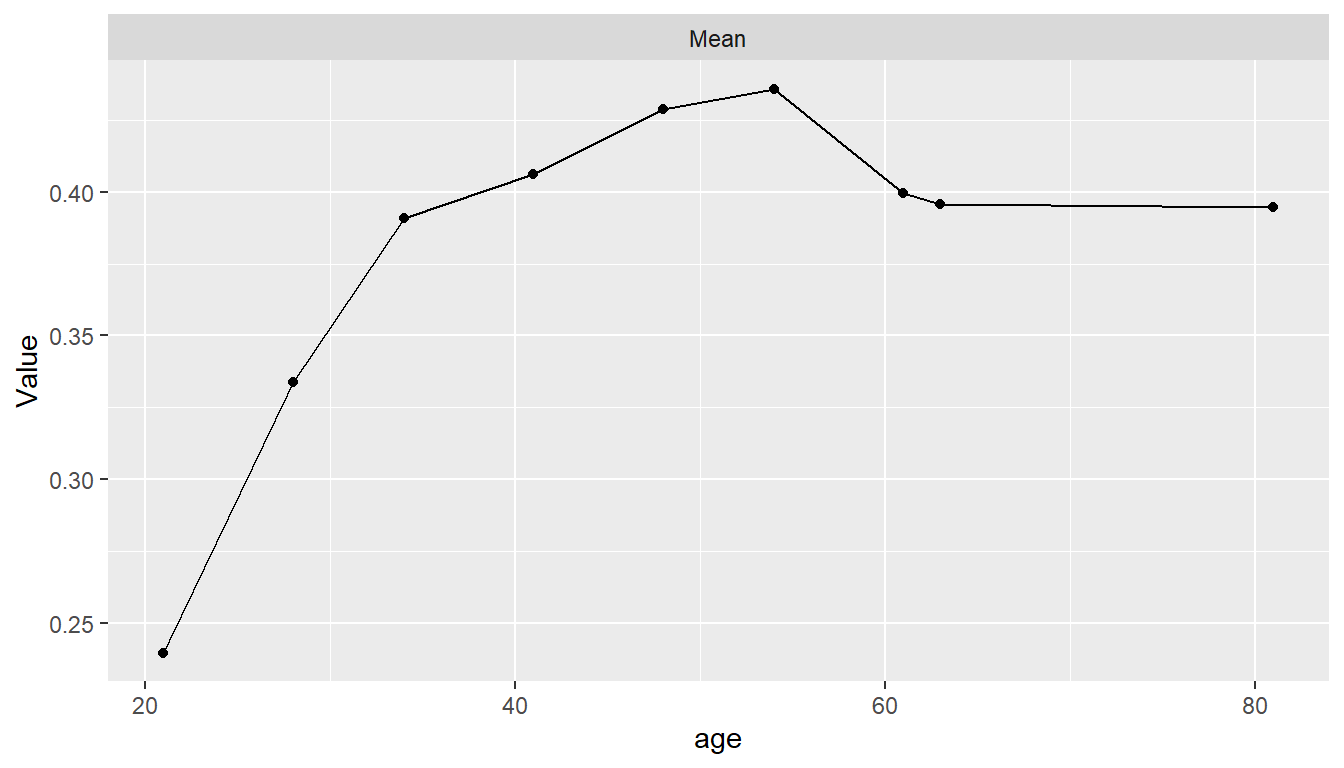
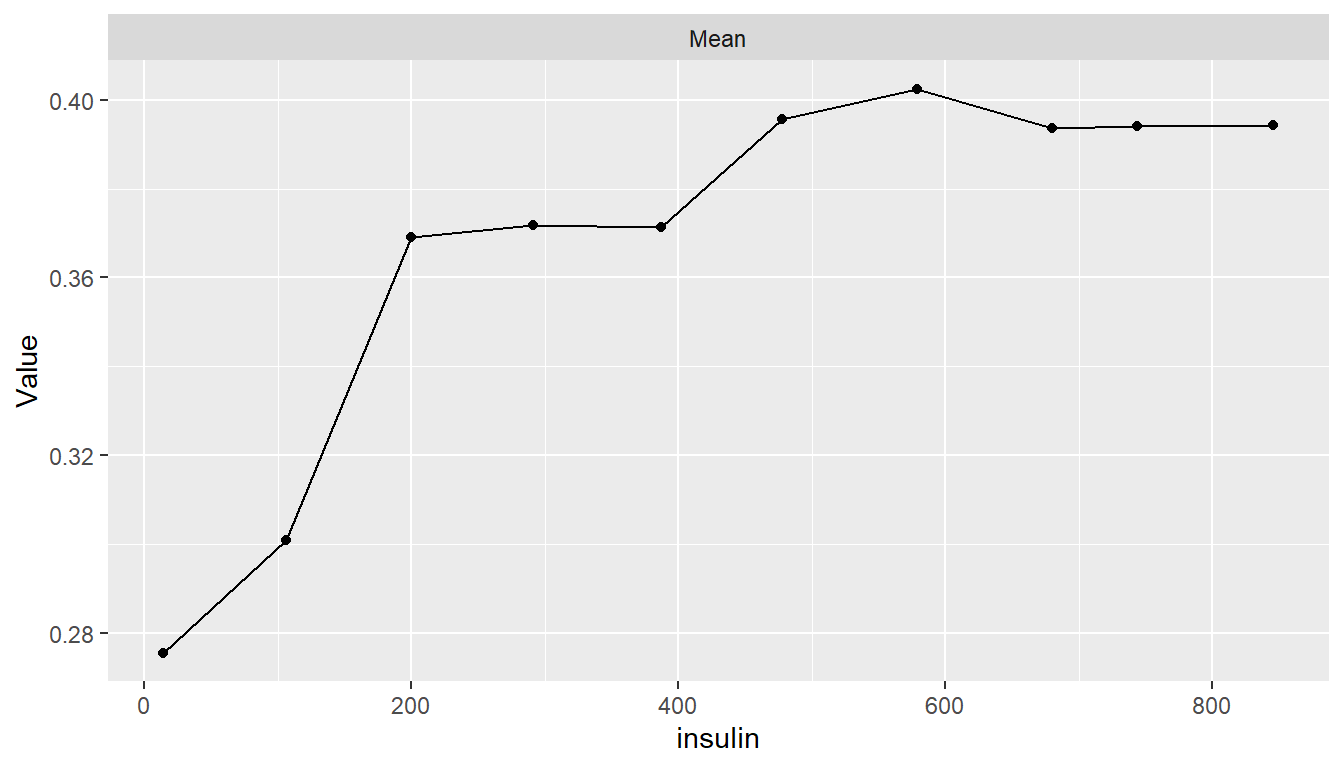
Partial Dependence Plots
pd <- dependence(model_fit, select = c(glucose, age, insulin))
plot(pd)